Uaa Uga And Uag Are All Codons
News Leon
Mar 25, 2025 · 7 min read
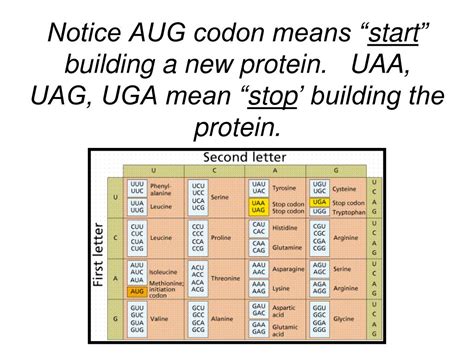
Table of Contents
UAA, UGA, and UAG: Understanding the Stop Codons
The genetic code, a fundamental concept in molecular biology, dictates how the sequence of nucleotides in DNA translates into the sequence of amino acids that make up proteins. This translation process is crucial for life, as proteins are the workhorses of cells, carrying out a vast array of functions. A key element of this code involves codons, three-nucleotide sequences that specify a particular amino acid. While most codons code for amino acids, three special codons – UAA, UGA, and UAG – stand out: these are the stop codons, also known as termination codons or nonsense codons. These codons signal the end of protein synthesis, marking the point at which the ribosome, the protein-synthesizing machinery of the cell, releases the newly synthesized polypeptide chain.
The Role of Stop Codons in Protein Synthesis
The process of protein synthesis, or translation, involves several key steps:
-
Initiation: The ribosome binds to the messenger RNA (mRNA) molecule at a specific initiation codon (usually AUG, which codes for methionine).
-
Elongation: The ribosome moves along the mRNA, reading each codon sequentially. For each codon, a corresponding transfer RNA (tRNA) molecule, carrying the appropriate amino acid, binds to the ribosome. The amino acids are then linked together to form a growing polypeptide chain.
-
Termination: When the ribosome encounters a stop codon (UAA, UGA, or UAG), it signals the end of the protein synthesis process. Release factors, proteins that recognize stop codons, bind to the ribosome, causing the release of the newly synthesized polypeptide chain. The ribosome then dissociates from the mRNA.
The accuracy of stop codon recognition is critical for producing functional proteins. Premature termination due to errors in recognition can result in truncated, non-functional proteins, potentially leading to various cellular malfunctions and diseases. Conversely, failure to recognize a stop codon results in a read-through of the mRNA sequence, potentially producing a protein with an extended, often non-functional, C-terminal tail.
Stop Codon Specificity and Release Factors
While all three stop codons signal the termination of translation, they aren't entirely interchangeable. The efficiency and specificity of stop codon recognition are influenced by the presence of specific release factors (RFs). These proteins bind to the ribosome at the A site (aminoacyl site) in response to a stop codon, catalyzing the hydrolysis of the peptidyl-tRNA bond and thus releasing the completed polypeptide.
In bacteria, two release factors are primarily responsible for stop codon recognition:
- RF1: Recognizes UAA and UAG.
- RF2: Recognizes UAA and UGA.
A third factor, RF3, acts as a GTPase, facilitating the release of RF1 and RF2 from the ribosome after termination.
Eukaryotes, on the other hand, utilize a single release factor, eRF1, which recognizes all three stop codons. A second factor, eRF3, is a GTPase similar to RF3 in bacteria, assisting in the recycling of eRF1.
The subtle differences in release factor recognition contribute to the observed variations in the frequency of usage of the different stop codons across different organisms and genes.
Stop Codon Usage Bias: Why the Differences?
The relative frequencies of UAA, UGA, and UAG vary significantly across different organisms and even within different genes of the same organism. This phenomenon, known as stop codon usage bias, is a complex issue influenced by several factors:
-
Codon context: The nucleotides surrounding the stop codon can influence the efficiency of termination. Specific nucleotide sequences might enhance or hinder the binding of release factors.
-
mRNA stability: The presence of a particular stop codon might influence the stability of the mRNA molecule, affecting the overall efficiency of translation.
-
tRNA availability: While stop codons don't directly interact with tRNAs, the availability of tRNAs for codons preceding the stop codon could indirectly affect termination efficiency. A bottleneck in tRNA availability could slow down the translation process and influence stop codon recognition.
-
Translational efficiency: Certain stop codons may be more efficiently recognized by the ribosome, leading to faster termination rates. This could be especially important in highly expressed genes where rapid protein synthesis is essential.
-
Evolutionary pressures: Over time, stop codon usage might be shaped by selective pressures, optimizing translation efficiency and minimizing errors in protein synthesis.
The precise mechanisms underlying stop codon usage bias are not fully understood, but research suggests that it plays a role in various cellular processes, including:
-
Protein folding: The choice of stop codon might influence the rate of protein folding and the final conformation of the protein.
-
Protein degradation: The location of the stop codon relative to specific protein degradation signals can affect the protein's half-life.
-
mRNA quality control: Stop codon usage could be linked to mechanisms ensuring the proper termination of translation and avoiding the production of aberrant proteins.
Stop Codon Mutations and Their Consequences
Mutations affecting stop codons can have significant consequences, leading to:
-
Nonsense mutations: These mutations convert a codon specifying an amino acid into a stop codon, resulting in premature termination of translation and the production of a truncated protein. These truncated proteins are often non-functional and can be detrimental to the cell. Nonsense mutations are implicated in various genetic diseases.
-
Readthrough mutations: These mutations, in contrast, convert a stop codon into a codon specifying an amino acid, leading to the extension of the protein beyond its normal length. This can also result in non-functional proteins and have deleterious effects on the cell.
-
Variations in stop codon usage: Subtle variations in stop codon usage, due to mutations in surrounding sequences, can subtly influence translation efficiency and protein function. These variations might contribute to phenotypic diversity or play a role in adaptation.
Clinical Significance of Stop Codons and Mutations
The significance of stop codons extends beyond the realm of basic research. Understanding their function and the implications of mutations affecting them is crucial in various areas of medicine:
-
Genetic disorders: Many genetic diseases arise from mutations that alter stop codons, leading to truncated or extended proteins that lose their function. Examples include cystic fibrosis, Duchenne muscular dystrophy, and various forms of inherited cancers.
-
Drug development: Drugs targeting the translation machinery, including those aimed at modulating stop codon recognition, are being developed to treat genetic disorders and cancers.
-
Diagnostics: The identification of stop codon mutations through genetic testing plays a critical role in the diagnosis of various genetic diseases.
-
Gene therapy: Strategies to correct stop codon mutations, such as nonsense-mediated mRNA decay inhibitors, are under investigation as potential gene therapy approaches.
Future Research Directions
Ongoing research continues to explore the intricacies of stop codon function and their influence on gene expression and cellular processes. Key areas of focus include:
-
Understanding the detailed mechanisms of stop codon recognition: Further investigations are needed to fully elucidate the interactions between release factors and the ribosome during termination.
-
Deciphering the complexities of stop codon usage bias: Research is ongoing to unravel the precise factors that contribute to the varying frequencies of stop codon usage across different organisms and genes.
-
Developing novel therapeutic strategies targeting stop codon mutations: Scientists are actively pursuing new approaches to correct or bypass stop codon mutations in order to restore protein function and treat genetic diseases.
Conclusion: UAA, UGA, and UAG – More Than Just the End of the Story
UAA, UGA, and UAG, while appearing as simple three-nucleotide sequences, play a vital and multifaceted role in the regulation of protein synthesis. Their accurate recognition is essential for producing functional proteins, and disruptions in this process can have profound consequences. Understanding these stop codons, their usage patterns, and the implications of mutations affecting them is crucial for advancing our knowledge of molecular biology, genetics, and medicine. Ongoing research continues to unravel the intricacies of their function, promising new insights and therapeutic strategies in the years to come. Their role is not simply to mark the end of a protein's synthesis, but also to influence aspects of protein function, mRNA stability, and ultimately, cellular processes and organismal health.
Latest Posts
Latest Posts
-
Reaction Of Ethanol And Acetic Acid
Mar 26, 2025
-
Proverbs Of The Day With Meaning
Mar 26, 2025
-
How Many Atoms Are In Sodium
Mar 26, 2025
-
The Urinary Bladder Is Composed Of What Epithelium
Mar 26, 2025
-
Sphere Is To Circle As Cube Is To
Mar 26, 2025
Related Post
Thank you for visiting our website which covers about Uaa Uga And Uag Are All Codons . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
