Okazaki Fragments Are Joined Together By
News Leon
Mar 19, 2025 · 7 min read
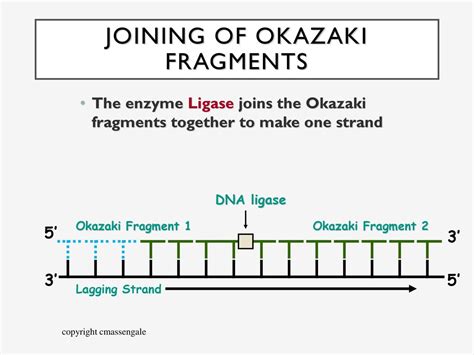
Table of Contents
Okazaki Fragments are Joined Together By: A Deep Dive into DNA Replication
DNA replication, the fundamental process by which life perpetuates itself, is a marvel of biological engineering. This intricate process involves the precise copying of a DNA molecule to create two identical daughter strands. While the leading strand replicates smoothly and continuously, the lagging strand presents a unique challenge. This challenge is overcome through the creation and subsequent joining of short DNA fragments known as Okazaki fragments. This article explores the detailed mechanism of how these fragments are joined together, delving into the essential enzymes and processes involved.
Understanding the Lagging Strand Problem
DNA polymerase, the enzyme responsible for synthesizing new DNA strands, can only add nucleotides to the 3' hydroxyl end of a pre-existing strand. This inherent directionality of DNA synthesis poses a problem for the lagging strand, which runs 5' to 3' in the direction of replication fork movement. Because of this, the lagging strand must be synthesized discontinuously in short, backward-moving segments – the Okazaki fragments.
The Role of RNA Primers
The initiation of each Okazaki fragment requires an RNA primer, synthesized by the enzyme primase. Primase is a type of RNA polymerase that doesn't require a pre-existing 3' hydroxyl group to start synthesis. These RNA primers provide the necessary 3' hydroxyl end for DNA polymerase to begin adding nucleotides to the lagging strand. The RNA primer is crucial because it creates the starting point for the Okazaki fragment synthesis. Without it, DNA polymerase would be unable to begin synthesizing the new DNA strand.
Joining Okazaki Fragments: A Multi-Step Process
The joining of Okazaki fragments is a complex, multi-step process involving several key enzymes:
1. DNA Polymerase I: Removing the RNA Primer
Once an Okazaki fragment has been synthesized, the RNA primer must be removed to ensure that the newly synthesized DNA strand is entirely composed of DNA. This crucial step is carried out by DNA polymerase I (Pol I). Pol I possesses both 5' to 3' exonuclease activity and 5' to 3' polymerase activity. Its 5' to 3' exonuclease activity allows it to remove the RNA nucleotides of the primer, while its polymerase activity simultaneously fills in the gaps with DNA nucleotides. This ensures a seamless transition from one Okazaki fragment to the next. The exonuclease activity is specifically targeted at the RNA primer; it doesn't degrade the newly synthesized DNA. This precise removal and replacement ensures accuracy in replication.
2. DNA Ligase: Sealing the Nick
After DNA polymerase I has removed the RNA primer and replaced it with DNA, a small nick or gap remains between the newly synthesized DNA and the preceding Okazaki fragment. This nick represents a discontinuity in the phosphodiester backbone of the DNA strand. This nick is then sealed by DNA ligase.
DNA ligase catalyzes the formation of a phosphodiester bond between the 3' hydroxyl group of one Okazaki fragment and the 5' phosphate group of the adjacent fragment. This reaction requires energy, usually provided by ATP or NAD+. The ligation process essentially "glues" the Okazaki fragments together, creating a continuous lagging strand. The precise and efficient action of DNA ligase ensures the integrity of the newly synthesized DNA molecule, preventing gaps or breaks that could compromise genetic information. The efficiency of this process is crucial for maintaining genome stability.
3. The Role of Sliding Clamps
Sliding clamps, such as the PCNA (proliferating cell nuclear antigen) in eukaryotes and the β-clamp in prokaryotes, play a crucial role in increasing the processivity of DNA polymerases. These ring-shaped proteins encircle the DNA and bind to DNA polymerases. This enhances the ability of DNA polymerases to synthesize long stretches of DNA without dissociating from the template. This is particularly important for DNA polymerase III during Okazaki fragment synthesis on the lagging strand, as it allows for efficient and rapid synthesis of multiple fragments. By increasing the processivity of DNA polymerase, sliding clamps contribute to the speed and accuracy of DNA replication.
4. The Importance of the Replication Fork
The replication fork, the Y-shaped structure where DNA replication occurs, is a dynamic region that coordinates the action of numerous enzymes and proteins. The coordinated action of helicases, single-strand binding proteins, primase, and DNA polymerases ensures that the leading and lagging strands are replicated efficiently and accurately. The structure of the replication fork plays an important role in the spatial arrangement of the enzymes involved in Okazaki fragment processing.
The precise choreography of these enzymes at the replication fork ensures that Okazaki fragments are synthesized, processed, and joined correctly, maintaining the integrity and stability of the genome. Any disruption to the activities of these enzymes or the structure of the replication fork can have potentially devastating consequences, leading to mutations or genomic instability.
Differences in Okazaki Fragment Processing Across Domains
While the fundamental process of Okazaki fragment joining is conserved across all domains of life (Bacteria, Archaea, and Eukarya), there are some subtle differences in the specifics of the enzymes and processes involved.
Prokaryotic Okazaki Fragment Processing
In prokaryotes, like E. coli, DNA polymerase III is the main enzyme responsible for synthesizing both leading and lagging strands. DNA polymerase I removes the RNA primers, and DNA ligase seals the nicks. The process is relatively straightforward and highly efficient, reflecting the faster replication rates observed in prokaryotic organisms.
Eukaryotic Okazaki Fragment Processing
In eukaryotes, the process is more complex, involving multiple DNA polymerases with specialized functions. The lagging strand synthesis is primarily carried out by DNA polymerase α and δ. DNA polymerase α initiates Okazaki fragment synthesis by creating a short RNA-DNA primer. DNA polymerase δ then extends this primer, synthesizing the majority of the DNA in the fragment. Flap endonuclease 1 (FEN1) removes the RNA portion of the primer, and DNA ligase I seals the nicks between adjacent fragments. The increased complexity likely reflects the larger genome size and the greater need for accuracy in eukaryotic DNA replication.
Errors in Okazaki Fragment Processing and Their Consequences
Errors in Okazaki fragment processing can lead to various problems, including:
- Mutations: Improper removal of RNA primers or inaccurate ligation can result in mutations, potentially causing genetic diseases or contributing to cancer development.
- Genome Instability: Unrepaired nicks or gaps in the DNA can lead to genomic instability, increasing the risk of chromosomal rearrangements and other deleterious events.
- Replication Fork Collapse: Problems with Okazaki fragment processing can cause the replication fork to stall or collapse, leading to DNA damage and cell death.
Clinical Significance and Research Directions
The understanding of Okazaki fragment processing is crucial for developing therapies for various diseases. For example, deficiencies in DNA polymerase I or DNA ligase can lead to various genetic disorders. Research focuses on understanding the precise mechanisms of Okazaki fragment processing, identifying new players involved, and exploring therapeutic targets for diseases related to defects in this crucial process. The continued study of Okazaki fragment processing promises to yield further insights into DNA replication and its relationship with human health.
Conclusion: A Precise and Essential Process
The joining of Okazaki fragments is a critical step in DNA replication. The intricate coordination of various enzymes, such as DNA polymerase I, DNA ligase, and primases, ensures the accurate and efficient synthesis of the lagging strand. This highly coordinated process maintains genome integrity and ensures the faithful transmission of genetic information from one generation to the next. Understanding the intricacies of Okazaki fragment processing is paramount for comprehending the fundamental processes of life and tackling the challenges posed by genetic diseases. Future research into the nuances of this process will continue to illuminate our understanding of DNA replication and its vital role in the preservation of life. Further investigation into the regulatory mechanisms controlling these enzymes and their interactions promises to unveil even more about this fascinating area of molecular biology. The precision and efficiency of Okazaki fragment processing serve as a testament to the elegance and complexity of biological systems.
Latest Posts
Latest Posts
-
How Many Neutrons Does Mg Have
Mar 19, 2025
-
The Material Used For Fuse Has Low Melting Point
Mar 19, 2025
-
In The Figure A Solid Sphere Of Radius
Mar 19, 2025
-
Where Is The Olecranon Process Found
Mar 19, 2025
-
The Joints Between Cranial Bones Of The Skull Are Called
Mar 19, 2025
Related Post
Thank you for visiting our website which covers about Okazaki Fragments Are Joined Together By . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
