Where Does Transcription Take Place In A Prokaryotic Cell
News Leon
Mar 20, 2025 · 7 min read
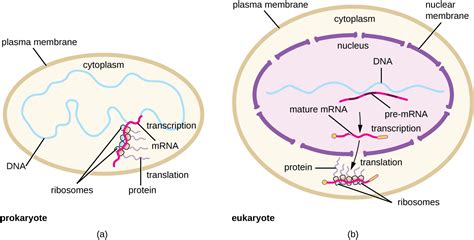
Table of Contents
Where Does Transcription Take Place in a Prokaryotic Cell?
Transcription, the crucial first step in gene expression, is a fascinating process with subtle yet significant differences between prokaryotic and eukaryotic cells. Understanding the location and mechanics of prokaryotic transcription is fundamental to grasping the simplicity and efficiency of gene regulation in these organisms. This article delves deep into the intricacies of prokaryotic transcription, exploring the cellular location, the players involved, and the key steps that lead to the synthesis of mRNA.
The Cytoplasm: The Central Hub of Prokaryotic Transcription
Unlike eukaryotes, where transcription occurs within the membrane-bound nucleus, prokaryotic transcription takes place in the cytoplasm. This seemingly simple difference has profound implications for gene regulation and the overall speed of gene expression. Because the processes of transcription and translation are not spatially separated, they are often coupled in prokaryotes. This means that translation of mRNA into protein can begin even before transcription is complete. This tightly coupled process allows for rapid responses to environmental changes and efficient utilization of resources.
Why the Cytoplasm? The Absence of a Nucleus
The absence of a membrane-bound nucleus is the defining characteristic that dictates the cytoplasmic location of prokaryotic transcription. The lack of this compartmentalization simplifies the process. There's no need to transport the newly synthesized mRNA across a nuclear membrane before translation can begin. This direct coupling of transcription and translation results in a much faster rate of protein synthesis compared to eukaryotes.
Key Players in Prokaryotic Transcription: A Molecular Orchestra
Several key molecular players orchestrate the intricate process of prokaryotic transcription. These include:
1. DNA: The Blueprint
The DNA molecule serves as the template for transcription. The specific sequence of DNA that codes for a particular RNA molecule is known as a gene. Prokaryotic genes are often organized into operons, clusters of genes under the control of a single promoter. This allows for coordinated regulation of multiple genes involved in a specific metabolic pathway. Understanding operons is crucial to fully grasping prokaryotic transcription.
2. RNA Polymerase: The Maestro
RNA polymerase is the central enzyme responsible for synthesizing RNA from a DNA template. Prokaryotes typically possess a single type of RNA polymerase, a complex enzyme composed of multiple subunits. This single enzyme is responsible for transcribing all types of RNA, including mRNA, tRNA, and rRNA.
Subunits of Prokaryotic RNA Polymerase: A Closer Look
The core enzyme of prokaryotic RNA polymerase consists of five subunits: two α (alpha), one β (beta), one β' (beta-prime), and one ω (omega). These subunits work together to assemble the RNA polymerase holoenzyme. The holoenzyme is formed when a sixth subunit, the sigma (σ) factor, binds to the core enzyme.
The sigma factor is absolutely crucial for the initiation of transcription. It's responsible for recognizing and binding to specific DNA sequences called promoters, located upstream of the gene. Different sigma factors can recognize different promoter sequences, allowing for the regulation of transcription under various environmental conditions. This is a vital aspect of prokaryotic adaptability.
3. Promoter Sequences: The Starting Signals
Promoter sequences are specific DNA sequences that signal the starting point for transcription. These sequences are recognized by the sigma factor of the RNA polymerase holoenzyme. The most commonly studied prokaryotic promoter sequences are the -10 and -35 regions, named for their positions relative to the transcription start site (+1). The consensus sequences for these regions are TATAAT (-10) and TTGACA (-35). Variations in these sequences can influence the strength of the promoter and thus the rate of transcription.
4. Ribosomes: The Translation Machinery
As mentioned earlier, in prokaryotes, translation begins before transcription is complete. This is due to the close proximity of ribosomes to the site of transcription. Ribosomes immediately bind to the nascent mRNA molecule, initiating translation of the mRNA into protein. This coupling of transcription and translation is a defining feature of prokaryotic gene expression.
The Three Stages of Prokaryotic Transcription: Initiation, Elongation, and Termination
Prokaryotic transcription unfolds in three distinct stages:
1. Initiation: Finding the Starting Point
Initiation begins with the binding of the RNA polymerase holoenzyme to the promoter region of the DNA. The sigma factor plays a critical role in this step, recognizing and binding to the -35 and -10 regions of the promoter. This binding leads to the unwinding of the DNA double helix, creating a transcription bubble. Once the transcription bubble is formed, RNA polymerase initiates RNA synthesis, using the template strand of DNA as a guide.
The initiation stage is a tightly regulated process, ensuring that transcription only begins at the appropriate time and in response to specific signals. The strength of the promoter, the availability of RNA polymerase, and the presence of regulatory proteins can all influence the rate of initiation.
2. Elongation: Building the RNA Chain
During elongation, RNA polymerase moves along the DNA template strand, unwinding the helix ahead of it and synthesizing a new RNA molecule complementary to the template strand. The RNA polymerase adds ribonucleotides to the 3' end of the growing RNA molecule, following the base-pairing rules (A with U, and G with C). The newly synthesized RNA molecule remains hybridized to the DNA template strand within the transcription bubble for a short distance, before separating and forming a double helix behind it. The elongation process continues until the RNA polymerase reaches a termination signal.
The accuracy of RNA synthesis is crucial, and errors during elongation can have significant consequences. RNA polymerase has a proofreading mechanism that helps to maintain fidelity during transcription, minimizing errors.
3. Termination: Ending the Process
Transcription termination signals the end of RNA synthesis. In prokaryotes, termination occurs in one of two primary ways:
-
Rho-independent termination: This type of termination involves specific DNA sequences in the newly synthesized RNA molecule that cause the RNA molecule to fold back on itself, forming a hairpin loop structure. This hairpin loop structure destabilizes the interaction between the RNA polymerase and the DNA template strand, leading to the termination of transcription. The sequence of the hairpin loop, rich in G-C base pairs, is crucial for the stability of this structure.
-
Rho-dependent termination: In this mechanism, a protein called Rho factor plays a crucial role. Rho factor is a hexameric ATPase that binds to specific sequences in the nascent RNA molecule and moves along the RNA, chasing the RNA polymerase. When Rho factor catches up to the RNA polymerase, it causes the release of both the RNA polymerase and the newly synthesized RNA molecule, thus terminating transcription.
Variations and Regulation: The Nuances of Prokaryotic Transcription
While the basic mechanism of prokaryotic transcription is relatively straightforward, there are numerous variations and levels of regulation that fine-tune gene expression to meet the specific needs of the organism. These include:
-
Operons: As previously mentioned, operons are clusters of genes transcribed as a single mRNA molecule. Operons allow for coordinated regulation of multiple genes involved in a common metabolic pathway. The lac operon in E. coli is a classic example, regulating genes involved in lactose metabolism.
-
Attenuation: This mechanism involves premature termination of transcription before the entire gene is transcribed. This process is often coupled with translation and is sensitive to the levels of specific metabolites.
-
Regulatory proteins: These proteins can bind to specific DNA sequences near the promoter, either enhancing or inhibiting transcription. These regulatory proteins can respond to various environmental signals, allowing for fine-tuned control of gene expression.
-
Sigma factors: Different sigma factors recognize different promoter sequences, enabling the bacterium to respond to various environmental changes by altering the expression of specific genes. For instance, heat shock sigma factors direct the expression of genes involved in stress responses.
Conclusion: A Dynamic and Efficient System
Prokaryotic transcription, occurring entirely within the cytoplasm, is a remarkably efficient and tightly regulated process. The close coupling of transcription and translation allows for rapid protein synthesis, enabling these organisms to respond quickly to environmental changes. The roles of RNA polymerase, sigma factors, promoter sequences, and termination mechanisms are finely balanced, orchestrating the precise expression of genes to maintain cellular function and survival. Understanding the complexities of prokaryotic transcription provides a foundation for advancing our knowledge of genetics, gene regulation, and the development of new therapeutic strategies targeting prokaryotic organisms. Further research continues to unveil the subtle nuances of this fascinating process and its critical role in the life of prokaryotes.
Latest Posts
Latest Posts
-
A Fully Loaded Slow Moving Freight Elevator
Mar 21, 2025
-
Why Is Hydrogen In Group 1
Mar 21, 2025
-
Adh Acts On Which Part Of Nephron
Mar 21, 2025
-
Which Of These Features Is A Characteristic Of Political Parties
Mar 21, 2025
-
There Are Two Forces On The 2 00 Kg Box
Mar 21, 2025
Related Post
Thank you for visiting our website which covers about Where Does Transcription Take Place In A Prokaryotic Cell . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
