Which Of The Following Enzymes Is Responsible For Rna Synthesis
News Leon
Mar 15, 2025 · 7 min read
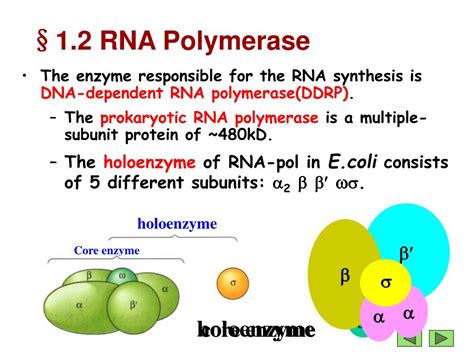
Table of Contents
Which of the Following Enzymes is Responsible for RNA Synthesis? Understanding RNA Polymerase
The central dogma of molecular biology dictates the flow of genetic information from DNA to RNA to protein. This process hinges on the precise and regulated synthesis of RNA molecules, a task carried out by a crucial enzyme family: RNA polymerases. This article delves deep into the world of RNA polymerases, exploring their structure, function, mechanisms, and the diverse roles they play in gene expression and cellular regulation. We'll also discuss the differences between RNA polymerases in prokaryotes and eukaryotes, highlighting their key characteristics and the implications for transcriptional control.
Understanding the Central Role of RNA Polymerase in RNA Synthesis
RNA synthesis, also known as transcription, is the process of creating an RNA molecule complementary to a DNA template strand. This process is fundamental to gene expression, as the synthesized RNA molecules serve various critical roles, including:
- Messenger RNA (mRNA): Carries genetic information from DNA to ribosomes, where it is translated into proteins.
- Transfer RNA (tRNA): Transports amino acids to the ribosome during protein synthesis.
- Ribosomal RNA (rRNA): Forms the structural and catalytic core of ribosomes.
- Small nuclear RNA (snRNA): Involved in splicing pre-mRNA molecules.
- MicroRNA (miRNA): Regulates gene expression by binding to mRNA molecules and inhibiting their translation.
RNA polymerase is the enzyme responsible for catalyzing the synthesis of all these RNA molecules. It achieves this by adding ribonucleotides to the 3' end of a growing RNA chain, using a DNA strand as a template. This process is highly regulated, ensuring that only the appropriate genes are expressed at the right time and in the right place.
The Structure and Function of RNA Polymerase
RNA polymerases are complex molecular machines, typically composed of multiple subunits. Their precise structure varies depending on the organism and the type of RNA being synthesized. However, some common features exist across different RNA polymerases:
- Active Site: This region contains the catalytic center responsible for the polymerization reaction. It binds to the incoming ribonucleotide triphosphates (NTPs) and catalyzes the formation of phosphodiester bonds between them.
- DNA Binding Domains: These regions allow the enzyme to bind specifically to the DNA template strand at the promoter region, a sequence upstream of the gene to be transcribed.
- Regulatory Domains: These domains interact with various transcription factors and other regulatory proteins that control the initiation, elongation, and termination of transcription.
Prokaryotic RNA Polymerase
Prokaryotes, such as bacteria and archaea, typically possess a single type of RNA polymerase. This enzyme is a multi-subunit complex consisting of:
- Core Enzyme: Composed of five subunits: α₂ββ'ω. The α subunits are involved in assembly and interaction with regulatory proteins. The β and β' subunits form the catalytic core, while the ω subunit stabilizes the enzyme.
- Sigma (σ) Factor: This subunit is crucial for promoter recognition and initiation of transcription. Different sigma factors can recognize different promoters, allowing for the regulated expression of specific genes in response to environmental changes.
The prokaryotic RNA polymerase holoenzyme (core enzyme + σ factor) binds to the promoter region, unwinds the DNA double helix, and initiates RNA synthesis. Transcription proceeds until a termination signal is encountered, causing the enzyme to dissociate from the DNA template.
Eukaryotic RNA Polymerases
Eukaryotes, including plants, animals, and fungi, have three main nuclear RNA polymerases, each responsible for synthesizing different types of RNA:
- RNA Polymerase I: Located in the nucleolus, this enzyme transcribes ribosomal RNA genes (rRNA).
- RNA Polymerase II: Located in the nucleoplasm, this enzyme transcribes protein-coding genes (mRNA) and some small nuclear RNAs (snRNAs).
- RNA Polymerase III: Located in the nucleoplasm, this enzyme transcribes transfer RNA genes (tRNA), some small nuclear RNAs (snRNAs), and 5S ribosomal RNA genes.
Eukaryotic RNA polymerases are more complex than their prokaryotic counterparts, requiring numerous general transcription factors (GTFs) for promoter recognition and initiation. These GTFs bind to specific promoter sequences, such as the TATA box, and recruit the RNA polymerase to the transcription start site. The process is further regulated by various other transcription factors that can either enhance or repress transcription.
The Mechanism of RNA Synthesis: A Detailed Look
The RNA synthesis process can be broken down into three main stages: initiation, elongation, and termination.
Initiation: Finding the Starting Point
Initiation involves the binding of RNA polymerase to the promoter region of the DNA template. This process is highly regulated and varies considerably between prokaryotes and eukaryotes.
- Prokaryotes: The sigma factor within the RNA polymerase holoenzyme plays a critical role in recognizing and binding to the promoter. Once bound, the enzyme unwinds the DNA helix at the transcription start site, forming a transcription bubble. The first ribonucleotide is then added, initiating RNA synthesis.
- Eukaryotes: The process is much more complex, requiring the assembly of a pre-initiation complex (PIC) at the promoter. This complex includes RNA polymerase II, various general transcription factors (GTFs), and other regulatory proteins. The PIC unwinds the DNA helix, and RNA polymerase II initiates transcription.
Elongation: Building the RNA Chain
Elongation is the stage where the RNA polymerase moves along the DNA template, synthesizing the RNA molecule. The enzyme adds ribonucleotides to the 3' end of the growing RNA chain, following the base-pairing rules (A with U, G with C). The DNA helix is unwound ahead of the enzyme and rewound behind it. This process is highly processive, meaning the enzyme remains bound to the DNA for long stretches, allowing for rapid RNA synthesis.
Termination: Ending the Synthesis
Termination is the final stage of transcription, involving the release of the RNA polymerase from the DNA template and the newly synthesized RNA molecule.
- Prokaryotes: Termination can occur through two main mechanisms: rho-independent (intrinsic) termination and rho-dependent termination. Rho-independent termination involves the formation of a hairpin structure in the RNA molecule, causing the RNA polymerase to pause and dissociate. Rho-dependent termination requires the rho factor, a protein that binds to the RNA and helps to unwind the RNA-DNA hybrid, causing termination.
- Eukaryotes: Termination is more complex and involves the processing of the pre-mRNA molecule. This includes the addition of a 5' cap, splicing out introns, and the addition of a 3' poly(A) tail. These processes are crucial for mRNA stability, export from the nucleus, and translation.
RNA Polymerase: Variations and Specificities
The diversity of RNA molecules necessitates the existence of specialized RNA polymerases with different properties and regulations. We've touched upon the fundamental differences between prokaryotic and eukaryotic systems. However, even within these broad categories, further variations and specificities exist.
For example, specific RNA polymerases might exhibit:
- Promoter specificity: Different RNA polymerases may recognize and bind to different promoter sequences, leading to the selective transcription of specific genes.
- Sensitivity to inhibitors: Different RNA polymerases may exhibit varying sensitivities to specific inhibitors, providing a useful tool for studying their activity.
- Regulation by transcription factors: The activity of RNA polymerases is intricately regulated by various transcription factors, leading to fine-tuned control over gene expression.
The Significance of RNA Polymerase in Research and Medicine
The understanding of RNA polymerases is crucial for:
- Drug development: RNA polymerase is a prime target for antibacterial and antiviral drugs. Inhibitors of bacterial RNA polymerase are widely used as antibiotics, while inhibitors of viral RNA polymerases are being developed as antiviral agents.
- Gene therapy: Manipulating the activity of RNA polymerase can be used to modulate gene expression, potentially correcting genetic defects or treating diseases.
- Understanding gene regulation: Studying RNA polymerases and their interactions with regulatory proteins is essential for understanding the complex mechanisms that control gene expression. This knowledge is crucial for tackling diseases linked to misregulated gene expression.
Conclusion: RNA Polymerase - The Master Architect of Gene Expression
In conclusion, RNA polymerases are essential enzymes responsible for the synthesis of all types of RNA molecules. Their structure, function, and mechanisms of action are intricately regulated, ensuring the precise and timely expression of genes. Understanding the nuances of RNA polymerases in both prokaryotes and eukaryotes is critical to advancing our knowledge of molecular biology and developing innovative therapies for various diseases. Further research into the complexities of RNA polymerase regulation and its interactions with other cellular machinery will continue to unlock valuable insights into the intricate world of gene expression and cellular processes. The future of understanding and manipulating gene expression relies heavily on our continuing efforts to unravel the mysteries of this remarkable enzyme.
Latest Posts
Related Post
Thank you for visiting our website which covers about Which Of The Following Enzymes Is Responsible For Rna Synthesis . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
